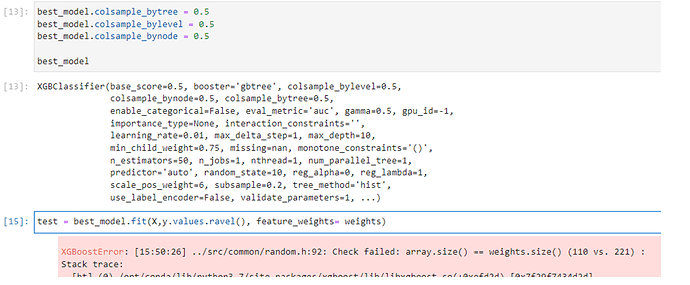
Hi , When I use feature_weights parameter , There is no different in the results…
1)not using feature_weights parameter
model5=xgb.XGBRegressor(objective=‘reg:squarederror’, colsample_bytree = 1, n_estimators =1000,gamma=0,subsample=1,reg_alpha=0.1,tree_method= “exact”)
model5.fit(train_x,train_y)
predictions=model5.predict(test_x)
2)using feature_wieghts parameter
model5=xgb.XGBRegressor(objective=‘reg:squarederror’, colsample_bytree = 1, n_estimators =1000,gamma=0,subsample=1,reg_alpha=0.1,tree_method= “exact”)
model5.fit(train_x,train_y,feature_weights=random_weights)
predictions=model5.predict(test_x)
The two cases have no differences.
I don’t know why ‘feature_weights’ doesn’t work… Please Help…