Hi,
I am using the sklearn python wrapper from xgboost 0.72.1 to train multiple boosted decision trees for a binary classification, all of them individually with early stopping, such that the best_ntree_limit differs.
When I use predict_proba on some data, I see that the ranges of the probabilities differ a lot, such that I am pretty sure the output does not correspond to a probability.
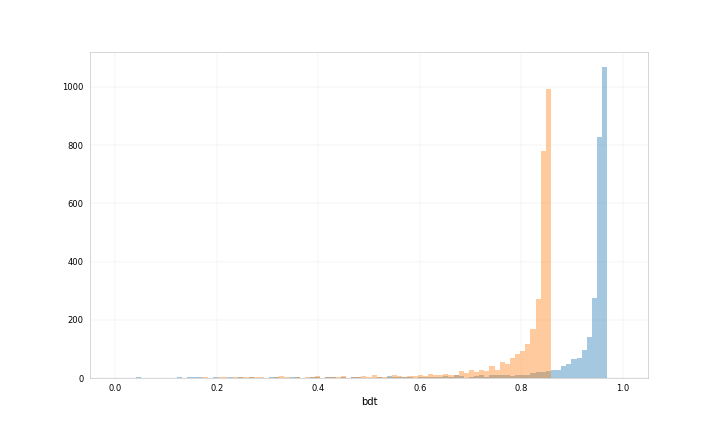
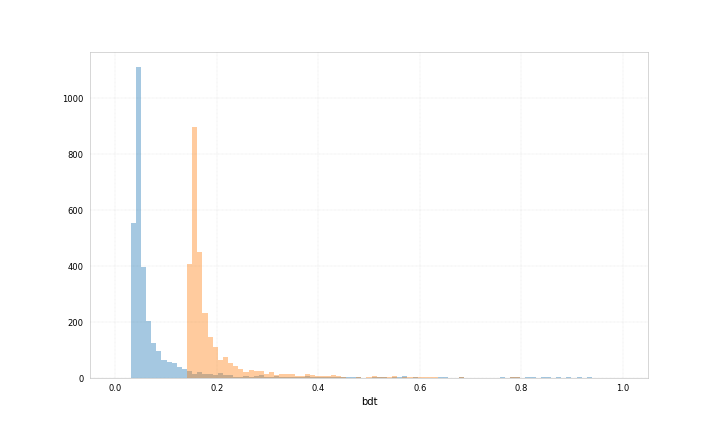
Here are example plots for data corresponding to category 0 and data corresponding to category 1, both showing the probability for the data to belong to category 1 predicted by two different bdts.
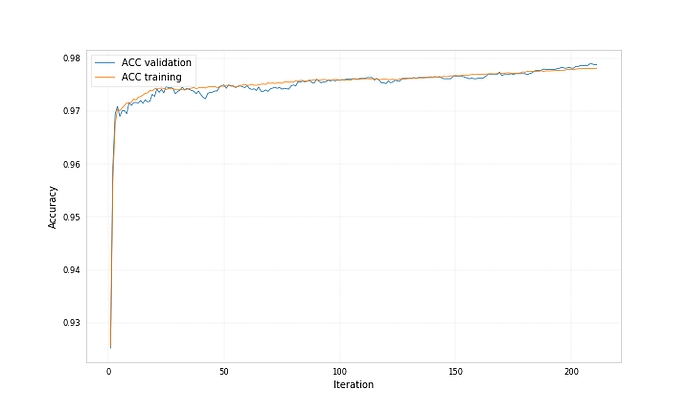
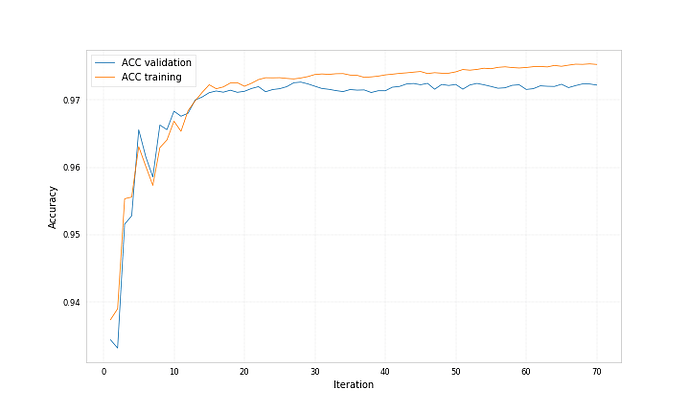
Both bdts were trained with ‘early_stopping_rounds’: 50, for one the best iteration is 212 for the other 71.
Is it possible, that the normalisation of the output breaks when not all trained trees are used to calculate it?
I saw that there was an issue in the git repo once (see https://github.com/dmlc/xgboost/issues/1897 ), where the solution (or error) was the existence of the key word argument output_margin, that you could pass to predict_proba. However this was removed in a more recent release, so I don’t know how to deal with this problem.
If the function name is predict_proba, I would naively expect the output to be a probability, so maybe its a bug?
Or am I making a mistake?
Many thanks in advance!