Hello,
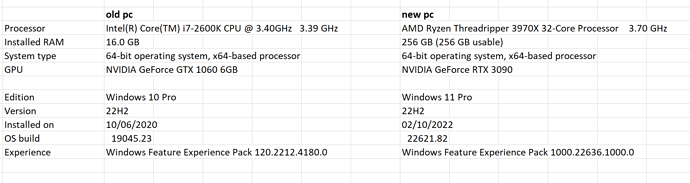
I have moved from a 12 year old pc with a GTX 1060 graphic card to a new pc with a RTX 3090 graphic card (see details below).
I was expecting a huge increase in speed. Although if I run a hyperparamtertuning-script in R on a dataset with 2 million observations and 110 features (from which 104 are from a one-hot-encoded factor) the processing time only halves.
To investigate if it could be caused by an improper R-installation, I have run a test-script (see below) on both R and Python on both my old and new pc.
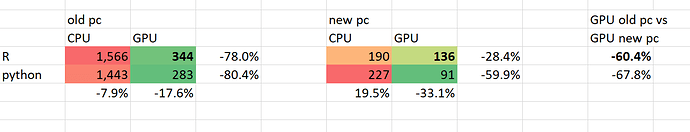
These are the processing times in seconds:
I am most interested in the improvement in speed for R since I am an R user. The improvement between the old and new pc for R on the GPU is 60.4%. For Python this is a approximate similar 67.8%. I conclude the R installation is fine.
Since the difference in configurations is quiet substantial, I was expecting a much larger improvement in speed. Is this justified or is a speed improvement of 60.4% as can be expected?
Thanks a lot!
Python script:
import pandas as pd
import numpy as np
import time
from sklearn.datasets import fetch_covtype
from sklearn.model_selection import train_test_split
import xgboost as xgb
# Fetch dataset using sklearn
cov = fetch_covtype()
X = cov.data
y = cov.target
# code snippet to save data as csv for use in R:
#df = pd.DataFrame(data=cov['data'], columns = cov['feature_names'])
#df.to_csv('my_cov.csv', sep = ',', index = False)
#np.savetxt("my_cov2.csv", y, delimiter=",")
# Create 0.75/0.25 train/test split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, train_size=0.75,
random_state=42)
# Specify sufficient boosting iterations to reach a minimum
num_round = 3000
# Leave most parameters as default
param = {'objective': 'multi:softmax', # Specify multiclass classification
'num_class': 8, # Number of possible output classes
'tree_method': 'gpu_hist' # Use GPU accelerated algorithm
}
# Convert input data from numpy to XGBoost format
dtrain = xgb.DMatrix(X_train, label=y_train)
dtest = xgb.DMatrix(X_test, label=y_test)
'''
gpu_res = {} # Store accuracy result
tmp = time.time()
# Train model
xgb.train(param, dtrain, num_round, evals=[(dtest, 'test')], evals_result=gpu_res)
print("GPU Training Time: %s seconds" % (str(time.time() - tmp)))
'''
# Repeat for CPU algorithm
tmp = time.time()
param['tree_method'] = 'hist'
cpu_res = {}
xgb.train(param, dtrain, num_round, evals=[(dtest, 'test')], evals_result=cpu_res)
print("CPU Training Time: %s seconds" % (str(time.time() - tmp)))
R script:
library(xgboost)
library(dplyr)
setwd("D:~/python_datafiles/")
# dataset laden:
mydata1 <- read.csv(file="my_cov.csv", header=TRUE, sep=",")
mydata2 <- read.csv(file="my_cov2.csv", header=FALSE, sep=",")
colnames(mydata2) <- "target"
mydata3 <- bind_cols(mydata1, mydata2)
mydata3$id <- 1:nrow(mydata3)
train <- mydata3 %>% dplyr::sample_frac(.75)
test <- dplyr::anti_join(mydata3, train, by = 'id')
train$id <- NULL
test$id <- NULL
val_targets <- test %>%
dplyr::select(target)
val_targets <- data.matrix(val_targets)
test$target <- NULL
val <- data.matrix(test)
xgb_val <- xgb.DMatrix(data = val, label = val_targets)
trainval_targets <- train %>%
dplyr::select(target)
trainval_targets <- data.matrix(trainval_targets)
train$target <- NULL
trainval <- data.matrix(train)
xgb_trainval <- xgb.DMatrix(data = trainval, label = trainval_targets)
start.time <- Sys.time()
model_n <- xgb.train(data = xgb_trainval,
tree_method = "hist",
objective = "multi:softmax",
num_class = 8,
nrounds = 3000,
print_every_n = 100,
watchlist = list(train = xgb_trainval, val = xgb_val)
)
end.time <- Sys.time()
time.taken <- end.time - start.time
time.taken